The direction of signal transmission is crucial for functions of neural networks. Therefore, the direction of signal flow, which is regulated by neuronal polarity, should be included when we analyze neural networks. However, the biochemical method used to identify neuronal polarity is time-consuming and may not be an appropriate strategy for analyzing large-scale neural networks.
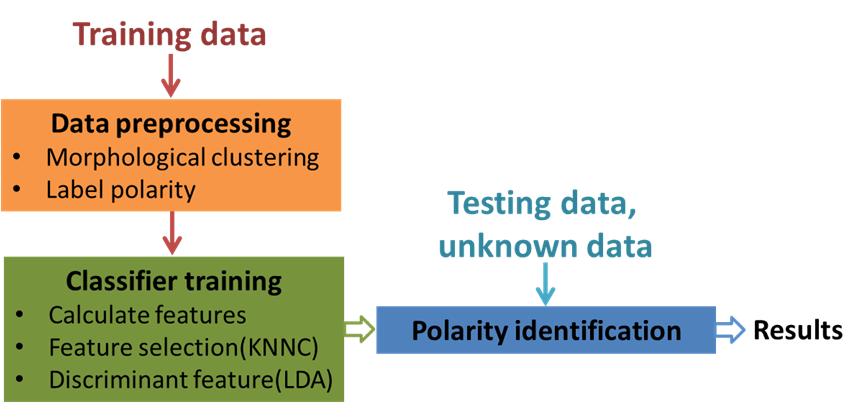
To address this problem, we proposed an algorithm for skeleton-based neuronal polarity identification (SPIN). In SPIN, we first morphologically divide a neuron into several substructures and for each substructure we extract seventeen morphological features. Next, K-nearest classifier (KNNC) is applied for identifying the most influential feature combination that correlates with the polarity of the training dataset. Finally, we perform linear discriminant analysis (LDA) to generate an optimal axis that provides highest accuracy for polarity identification. The optimal axis is then used to identify polarity of neurons in the testing set.
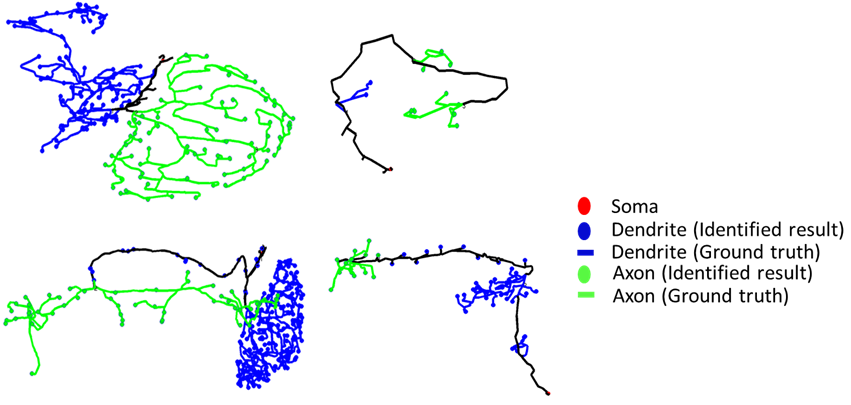
We tested this method on neurons innervating protocerebral bridge (PCB) or medulla (MED) in Drosophila. The neuron skeletons were extracted from data obtained from Brain Research Center, National Tsing Hua University, Taiwan. On average, the polarity of more than 85% terminal points in a neuron could be correctly identified. We tested the maximum performance of SPIN on a clean dataset constructed by manually removing artificial branches resulting from noise in raw images. We found that the average accuracy reaches 95% in the best case. Our results show that, as a computer-based semi-automatic procedure, SPIN provides quick polarity identification and is particularly suitable for analyzing large-scale data.